James D Munday, PhD

Infectious disease epidemiology and surveillance based in Basel
[ORCID] [Scholar] [bsky] [Mail]
Papers | Conferences | Posts
Wastewater based infectious disease surveillance and epidemiology
by jamesmunday
Background
Surveillance of pathogens spreading through a population is an essential tool in epidemic response and public health resource planning. Historically programes have used reported cases, hospitalisations and various sentinel systems to track infection prevalence and transmission in the population. For many infections, virus is shed in feces. In recent years protocols have been developed to quantify the concentration of viral RNA within sewage for a growing number of pathogens. By monitoring municipal wastewater systems it is possible to track fluctuations in prevalence of disease in the population served by the system.
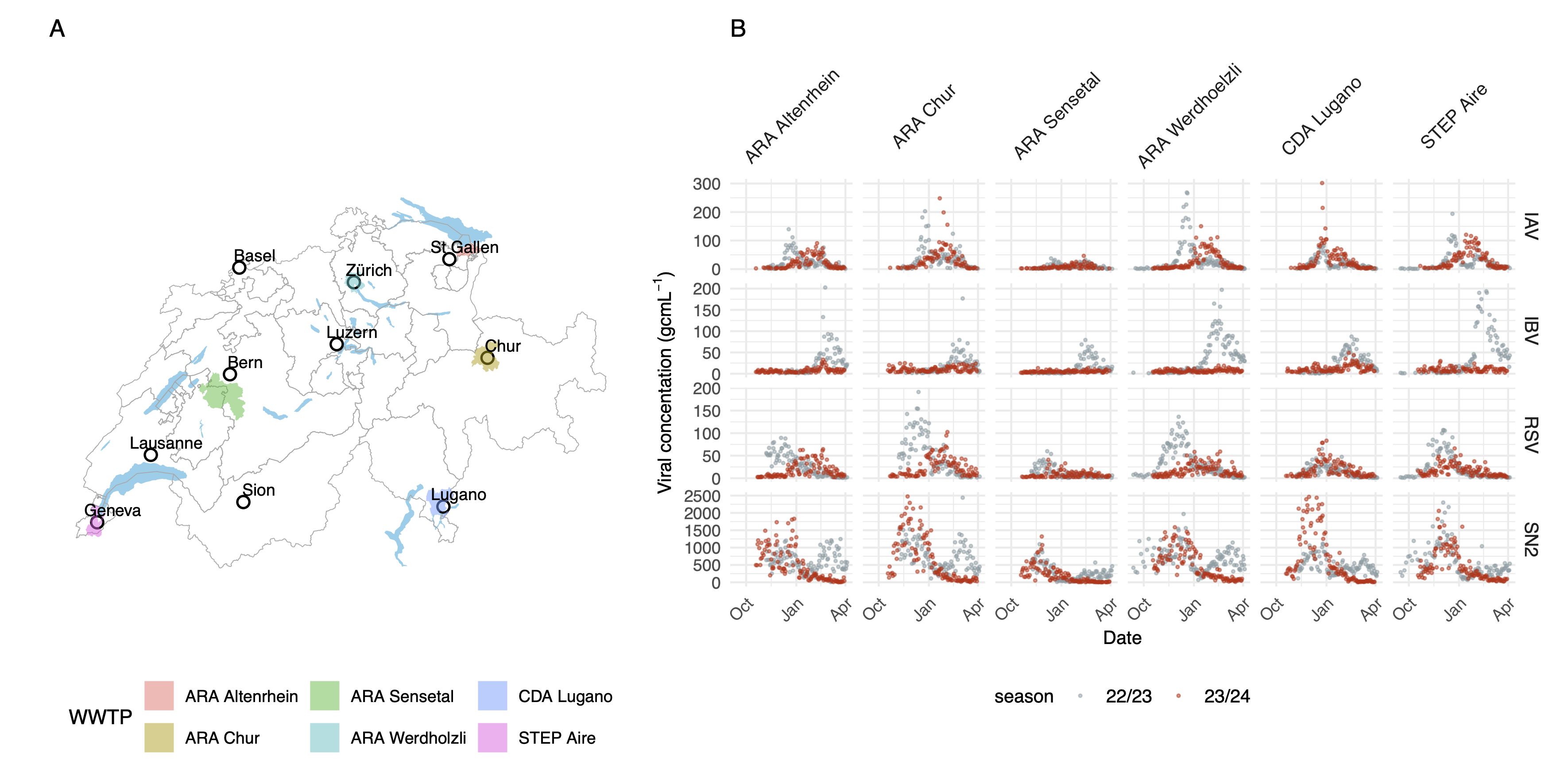
Wastewater treatment plant catchments in swizerland and viral concentrations measured in each of them for SARS-CoV-2 (SN2), Influenza A and B and RSV
Context
We are part of a collaboration in Switzerland that monitors wastewater from treatment plants across Switzerland. Samples are collected between 5 and 7 days a week, viral concentrations of SARS-CoV-2, Influenza A and B and RSV are quantified using digital deoplet PCR. Amplicons are also sequenced using illumina next generation sequncing.
Research
The focus of my research has been on how to optimise the collection of data and estimation of concentration to provide public health insights.
Firstly, with collaborators at Eawag, UniSante and FOPH we have evaluated the ability of different sampling strategies to inform trends in SARS-CoV-2 prevalence across switzerland and how this translates to reported cases and hospitalisations.
Secondly, with collaborators from Eawag we investigate degredation of viral samples between sampling and testing - which can affect the quality of concentration estimates, highlighting the importance of testing samples promptly - especially when concentrations are low. Samples are collected most days, and are processed in batches on one or 2 days per week to quantify the RNA concentrations for our target pathogens. Between collection and processing samples are stored in a regrigerator at the treatment plant. We know that RNA degrades over time in the sewer, although regrigeration should slow the rate at which RNA degrades, it does not eliminate it. The time between collection and processing therefore provides an opportunity for the RNA to degrade further, potentially impacting concentration estimates. Using a bayesian time-series model we quantified decline in viral concentrations correllated with the delay between collection and processing of samples over the course of our national monitoring programme. The degradation appeared to be most consistent and of greatest magnitufe for Influenza A and RSV. There was also substantial variation in the reduction of concentrations between sampling sites, suggesting that site specific factors may play a role in degradation of RNA.
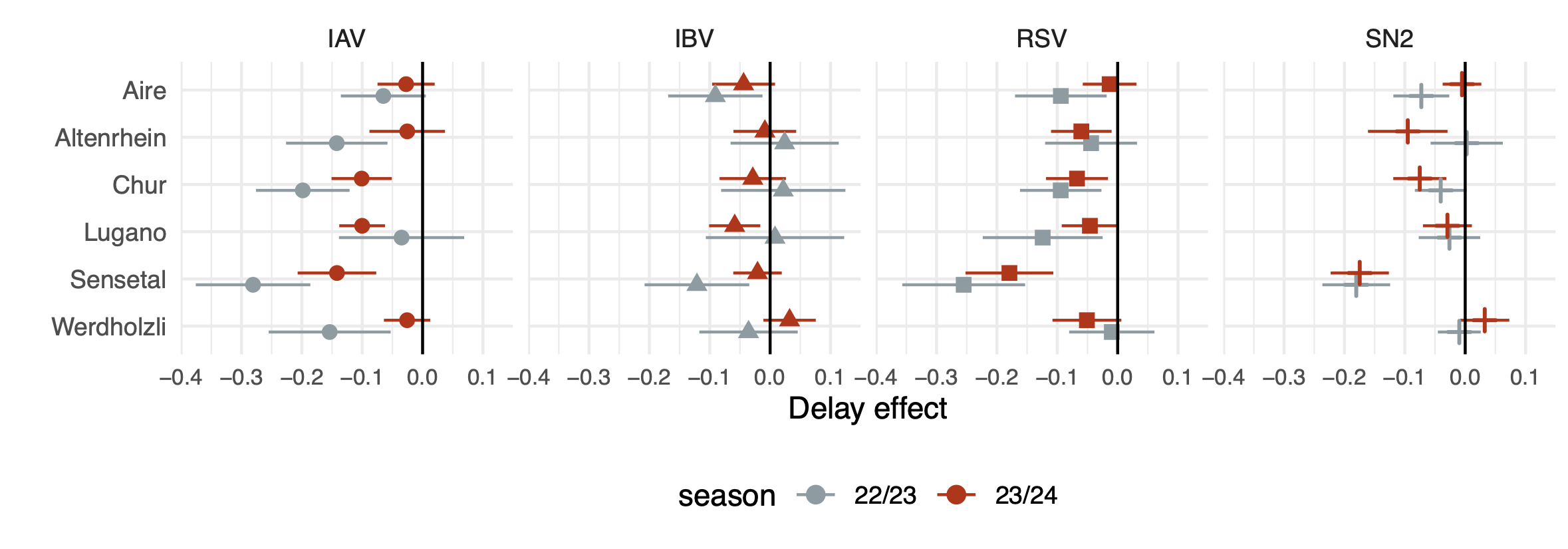
Inferred effect of delay between collection and processing on concentrations of SARS-CoV-2 (SN2), Influenza A and B and RSV RNA in wastewater from six treatment plants in switzerland during the 2022-2023 and 2023-2024 seasons.
We estimate that the reduction in viral concentrations associated with degradation between collection and sampling is sufficient that a substantial number of concentration estimates are brought below the “limit of detection”, a threshold where the probability of sample loss becomes unacceptably high.